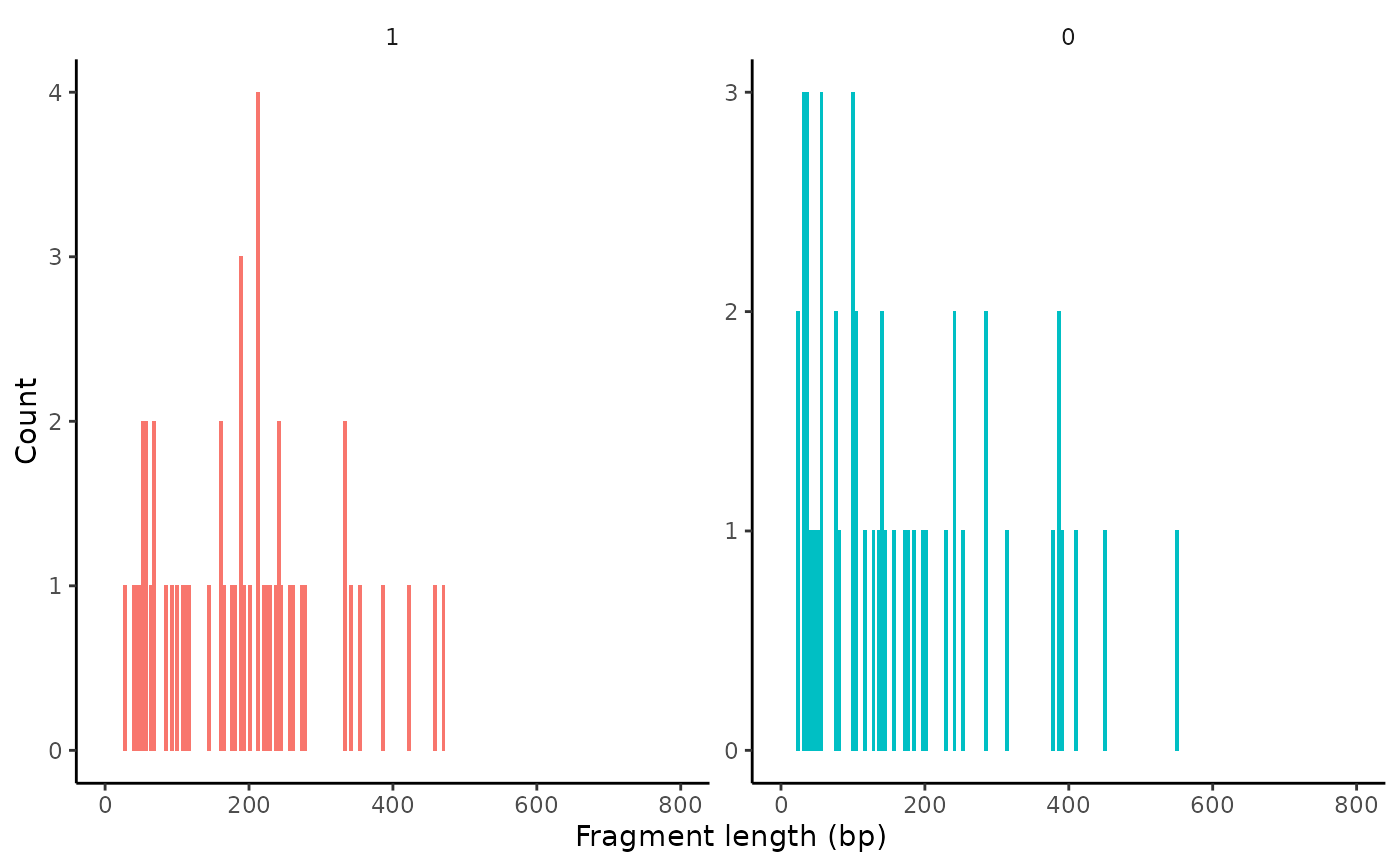
Plot the frequency that fragments of different lengths are present for different groups of cells.
FragmentHistogram( object, assay = NULL, region = "chr1-1-2000000", group.by = NULL, cells = NULL, log.scale = FALSE, ... )
Arguments
| object | A Seurat object |
|---|---|
| assay | Which assay to use. Default is the active assay. |
| region | Genomic range to use. Default is fist two megabases of chromosome 1. Can be a GRanges object, a string, or a vector of strings. |
| group.by | Name of one or more metadata columns to group (color) the cells by. Default is the current cell identities |
| cells | Which cells to plot. Default all cells |
| log.scale | Display Y-axis on log scale. Default is FALSE. |
| ... | Arguments passed to other functions |
Value
Returns a ggplot object
Examples
# \donttest{ fpath <- system.file("extdata", "fragments.tsv.gz", package="Signac") Fragments(atac_small) <- CreateFragmentObject( path = fpath, cells = colnames(atac_small), validate.fragments = FALSE )#>FragmentHistogram(object = atac_small, region = "chr1-10245-780007")#> Warning: Removed 4 rows containing missing values (geom_bar).# }