Parallel and distributed processing
Compiled: April 28, 2021
Source:vignettes/future.Rmd
future.RmdParallel computing is supported in Signac through the future package, making it easy to specify different parallelization options. Here we demonstrate parallelization of the FeatureMatrix function and show some benchmark results to get a sense for the amount of speedup you might expect.
The Seurat package also uses future for parallelization, and you can see the Seurat vignette for more information.
How to enable parallelization in Signac
Parallelization can be enabled simply by importing the future package and setting the plan.
## sequential:
## - args: function (..., envir = parent.frame())
## - tweaked: FALSE
## - call: NULLBy default the plan is set to sequential processing (no parallelization). We can change this to multicore, multiprocessor, or multisession to get asynchronous processing, and set the number of workers to change the number of cores used.
## multiprocess:
## - args: function (..., envir = parent.frame(), workers = 10)
## - tweaked: TRUE
## - call: plan("multiprocess", workers = 10)You might also need to increase the maximum memory usage:
options(future.globals.maxSize = 50 * 1024 ^ 3) # for 50 Gb RAMNote that as of future version 1.14.0, forked processing is disabled when running in RStudio. To enable parallel computing in RStudio, you will need to select the “multisession” option.
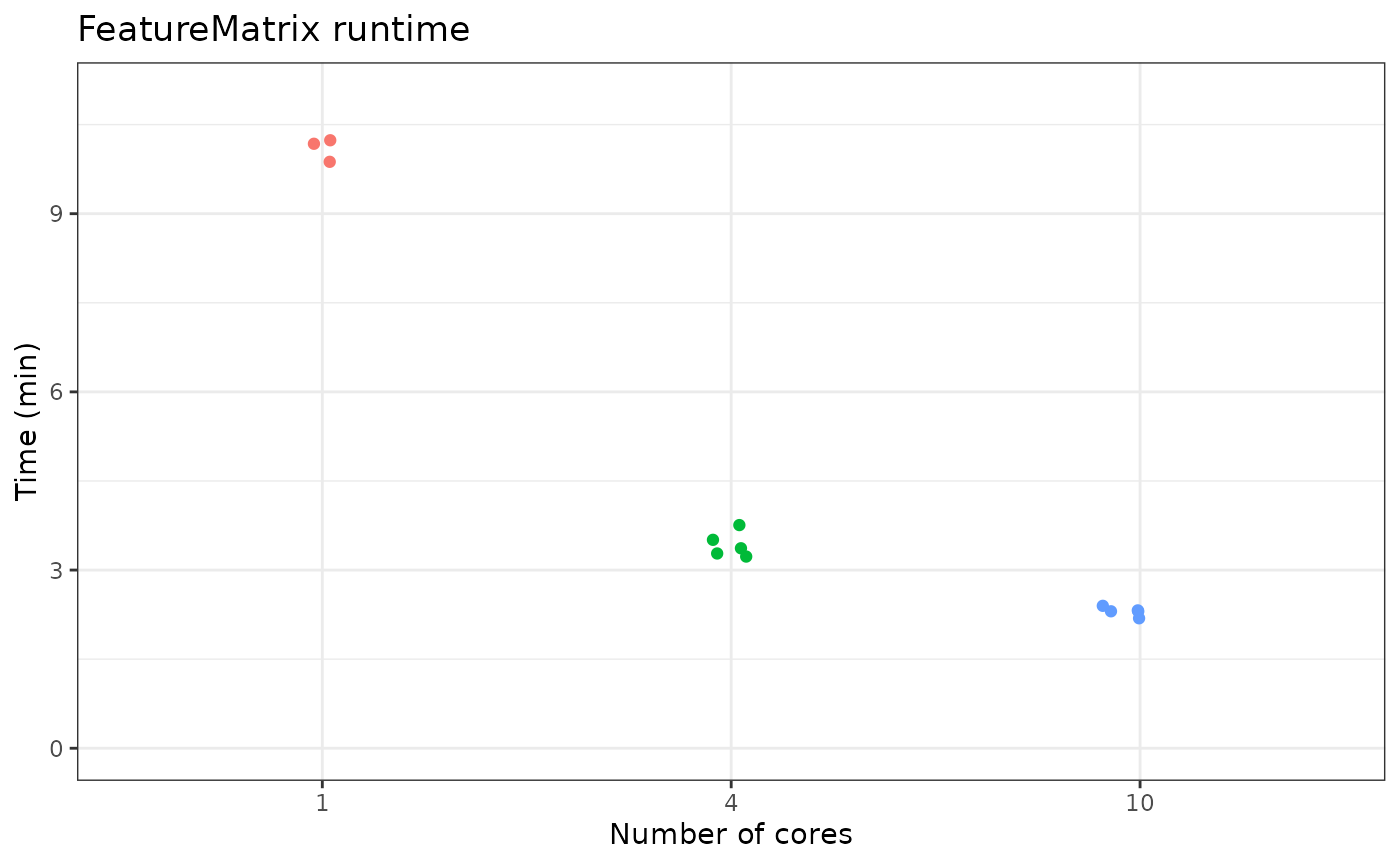
Benchmarking
Here we demonstrate the runtime of FeatureMatrix run on 90,686 peaks for 10,247 human PBMCs under different parallelization options:
View benchmarking code
The following code was run on Ubuntu 18.04 LTS with 12 Intel Xeon W-2135 CPUs @ 3.70GHz and 500 Gb RAM.
library(Signac)
# load data
fragments <- "/home/stuartt/data/pbmc10k/atac_pbmc_10k_nextgem_fragments.tsv.gz"
peaks.10k <- read.table(
file = "/home/stuartt/data/pbmc10k/atac_pbmc_10k_nextgem_peaks.bed",
col.names = c("chr", "start", "end")
)
peaks <- GenomicRanges::makeGRangesFromDataFrame(peaks.10k)
cells <- readLines("/home/stuartt/data/pbmc10k/cells.txt")
fragments <- CreateFragmentObject(path = fragments, cells = cells, validate.fragments = FALSE)
# set number of replicates
nrep <- 5
results <- data.frame()
process_n <- 2000
# run sequentially
timing.sequential <- c()
for (i in seq_len(nrep)) {
start <- Sys.time()
fmat <- FeatureMatrix(fragments = fragments, features = peaks, cells = cells, process_n = process_n)
timing.sequential <- c(timing.sequential, as.numeric(Sys.time() - start, units = "secs"))
}
res <- data.frame(
"setting" = rep("Sequential", nrep),
"cores" = rep(1, nrep),
"replicate" = seq_len(nrep),
"time" = timing.sequential
)
results <- rbind(results, res)
# 4 core
library(future)
plan("multiprocess", workers = 4)
options(future.globals.maxSize = 100000 * 1024^2)
timing.4core <- c()
for (i in seq_len(nrep)) {
start <- Sys.time()
fmat <- FeatureMatrix(fragments = fragments, features = peaks, cells = cells, process_n = process_n)
timing.4core <- c(timing.4core, as.numeric(Sys.time() - start, units = "secs"))
}
res <- data.frame(
"setting" = rep("Parallel", nrep),
"cores" = rep(4, nrep),
"replicate" = seq_len(nrep),
"time" = timing.4core
)
results <- rbind(results, res)
# 10 core
plan("multiprocess", workers = 10)
timing.10core <- c()
for (i in seq_len(nrep)) {
start <- Sys.time()
fmat <- FeatureMatrix(fragments = fragments, features = peaks, cells = cells, process_n = process_n)
timing.10core <- c(timing.10core, as.numeric(Sys.time() - start, units = "secs"))
}
res <- data.frame(
"setting" = rep("Parallel", nrep),
"cores" = rep(10, nrep),
"replicate" = seq_len(nrep),
"time" = timing.10core
)
results <- rbind(results, res)
# save results
write.table(
x = results,
file = paste0("/home/stuartt/github/chrom/bldignore/timings_", Sys.date(), ".tsv"),
quote = FALSE,
row.names = FALSE
)
Session Info
## R version 4.0.1 (2020-06-06)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 18.04.5 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.7.1
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.7.1
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggplot2_3.3.3 future_1.21.0
##
## loaded via a namespace (and not attached):
## [1] highr_0.9 pillar_1.6.0 bslib_0.2.4 compiler_4.0.1
## [5] jquerylib_0.1.4 tools_4.0.1 digest_0.6.27 jsonlite_1.7.2
## [9] evaluate_0.14 memoise_2.0.0 lifecycle_1.0.0 tibble_3.1.1
## [13] gtable_0.3.0 pkgconfig_2.0.3 rlang_0.4.10 DBI_1.1.1
## [17] yaml_2.2.1 parallel_4.0.1 pkgdown_1.6.1 xfun_0.22
## [21] fastmap_1.1.0 withr_2.4.2 dplyr_1.0.5 stringr_1.4.0
## [25] knitr_1.33 generics_0.1.0 vctrs_0.3.7 desc_1.3.0
## [29] fs_1.5.0 sass_0.3.1 systemfonts_1.0.1 globals_0.14.0
## [33] tidyselect_1.1.0 rprojroot_2.0.2 grid_4.0.1 glue_1.4.2
## [37] listenv_0.8.0 R6_2.5.0 textshaping_0.3.3 fansi_0.4.2
## [41] parallelly_1.24.0 rmarkdown_2.7 farver_2.1.0 purrr_0.3.4
## [45] magrittr_2.0.1 scales_1.1.1 codetools_0.2-18 htmltools_0.5.1.1
## [49] ellipsis_0.3.1 assertthat_0.2.1 colorspace_2.0-0 labeling_0.4.2
## [53] ragg_1.1.2 utf8_1.2.1 stringi_1.5.3 munsell_0.5.0
## [57] cachem_1.0.4 crayon_1.4.1