Transcription factor footprinting
Compiled: October 11, 2025
Source:vignettes/footprint.Rmd
footprint.RmdData loading
For this vignette we’ll use the dataset introduced and pre-processed in the trajectory building vignette.
## Warning: `aes_string()` was deprecated in ggplot2 3.0.0.
## ℹ Please use tidy evaluation idioms with `aes()`.
## ℹ See also `vignette("ggplot2-in-packages")` for more information.
## ℹ The deprecated feature was likely used in the Seurat package.
## Please report the issue at <https://github.com/satijalab/seurat/issues>.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.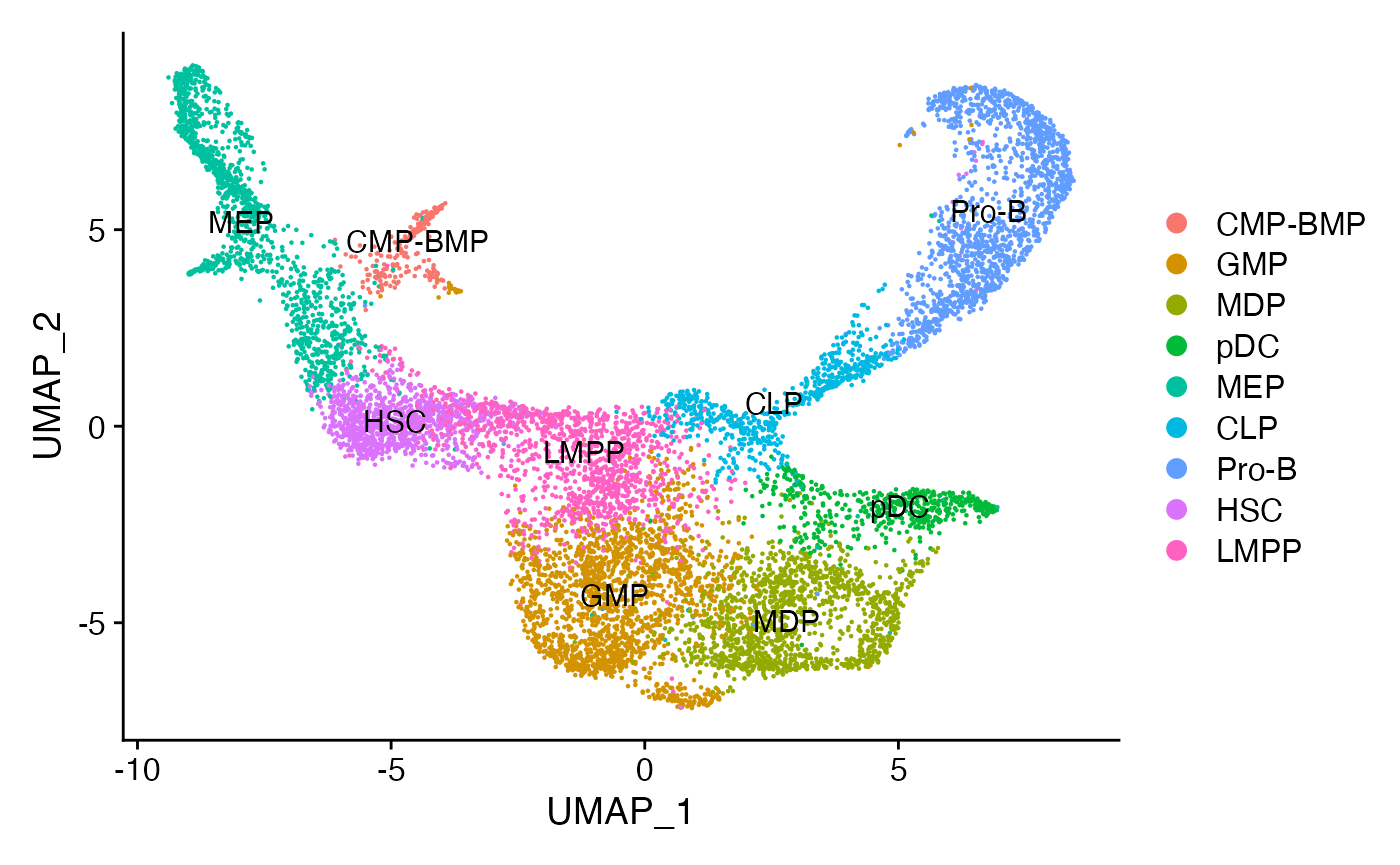
To perform a footprinting analysis we first need to add motif information to the object, including the exact positions of each motif. This can be done using functions from the and packages.
library(motifmatchr)
library(JASPAR2020)
library(TFBSTools)
library(BSgenome.Hsapiens.UCSC.hg19)
# extract position frequency matrices for the motifs
pwm <- getMatrixSet(
x = JASPAR2020,
opts = list(species = 9606, all_versions = FALSE)
)
# add motif information
bone <- AddMotifs(bone, genome = BSgenome.Hsapiens.UCSC.hg19, pfm = pwm)Motif footprinting
Now we can footprint any motif that we have positional information
for. By default, this includes every instance of the motif in the
genome. We can instead use the in.peaks = TRUE parameter to
include only those motifs that fall inside a peak in the assay. The
Footprint() function gathers all the required data and
stores it in the assay. We can then plot the footprinted motifs using
the PlotFootprint() function.
# gather the footprinting information for sets of motifs
bone <- Footprint(
object = bone,
motif.name = c("GATA2", "CEBPA", "EBF1"),
genome = BSgenome.Hsapiens.UCSC.hg19
)
# plot the footprint data for each group of cells
p2 <- PlotFootprint(bone, features = c("GATA2", "CEBPA", "EBF1"))p2 + patchwork::plot_layout(ncol = 1)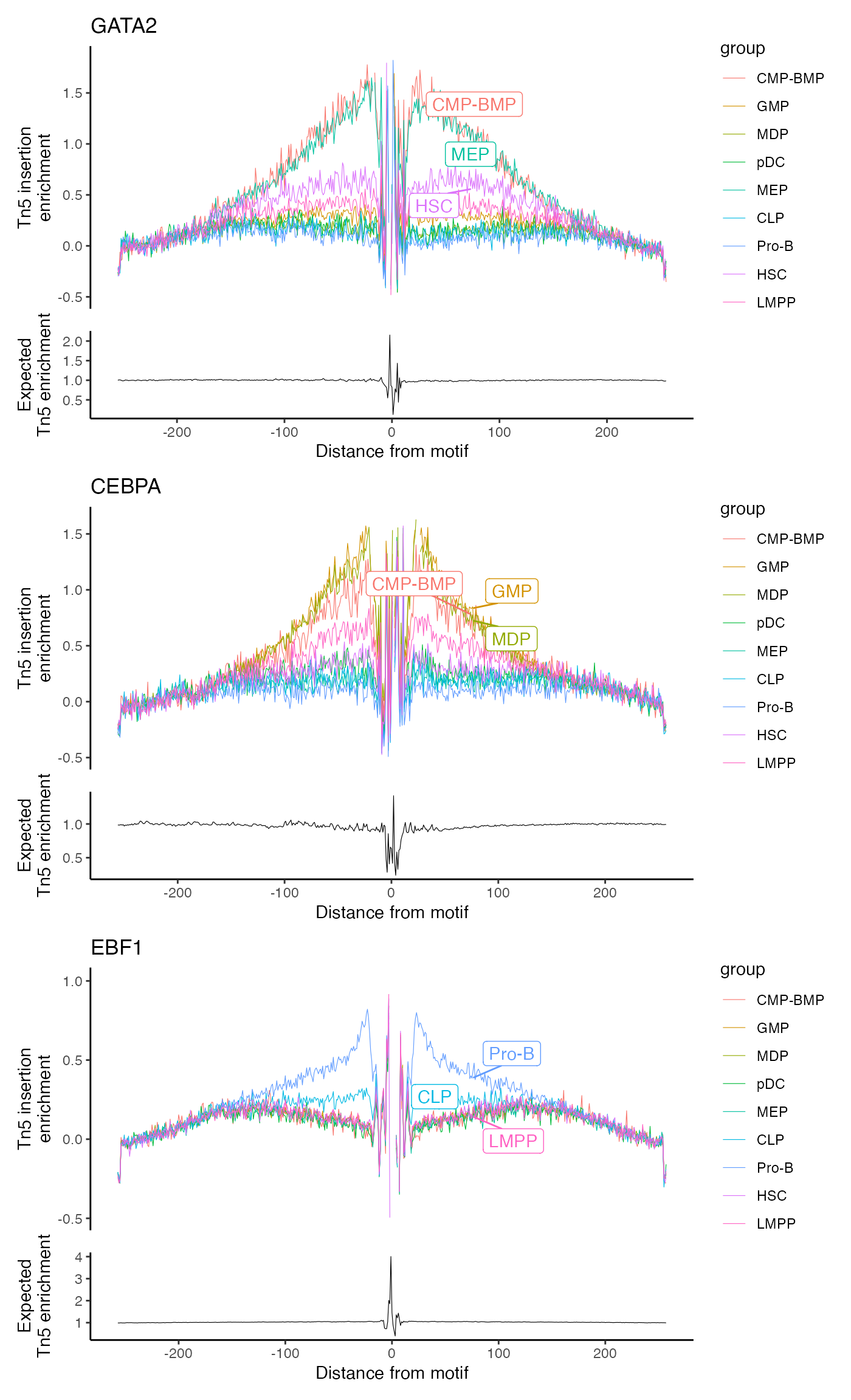
Session Info
## R version 4.5.1 (2025-06-13)
## Platform: aarch64-apple-darwin20
## Running under: macOS Tahoe 26.0.1
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.1
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: Asia/Singapore
## tzcode source: internal
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] BSgenome.Hsapiens.UCSC.hg19_1.4.3 BSgenome_1.76.0
## [3] rtracklayer_1.68.0 BiocIO_1.18.0
## [5] Biostrings_2.76.0 XVector_0.48.0
## [7] GenomicRanges_1.60.0 GenomeInfoDb_1.44.3
## [9] IRanges_2.42.0 S4Vectors_0.46.0
## [11] BiocGenerics_0.54.0 generics_0.1.4
## [13] TFBSTools_1.46.0 JASPAR2020_0.99.10
## [15] motifmatchr_1.30.0 Seurat_5.3.0
## [17] SeuratObject_5.2.0 sp_2.2-0
## [19] Signac_1.16.0
##
## loaded via a namespace (and not attached):
## [1] RcppAnnoy_0.0.22 splines_4.5.1
## [3] later_1.4.4 bitops_1.0-9
## [5] tibble_3.3.0 polyclip_1.10-7
## [7] XML_3.99-0.19 DirichletMultinomial_1.50.0
## [9] fastDummies_1.7.5 lifecycle_1.0.4
## [11] pwalign_1.4.0 globals_0.18.0
## [13] lattice_0.22-7 MASS_7.3-65
## [15] magrittr_2.0.4 plotly_4.11.0
## [17] sass_0.4.10 rmarkdown_2.30
## [19] jquerylib_0.1.4 yaml_2.3.10
## [21] httpuv_1.6.16 sctransform_0.4.2
## [23] spam_2.11-1 spatstat.sparse_3.1-0
## [25] reticulate_1.43.0 cowplot_1.2.0
## [27] pbapply_1.7-4 DBI_1.2.3
## [29] RColorBrewer_1.1-3 abind_1.4-8
## [31] Rtsne_0.17 purrr_1.1.0
## [33] RCurl_1.98-1.17 GenomeInfoDbData_1.2.14
## [35] ggrepel_0.9.6 irlba_2.3.5.1
## [37] listenv_0.9.1 spatstat.utils_3.2-0
## [39] seqLogo_1.74.0 goftest_1.2-3
## [41] RSpectra_0.16-2 spatstat.random_3.4-2
## [43] fitdistrplus_1.2-4 parallelly_1.45.1
## [45] pkgdown_2.1.3 DelayedArray_0.34.1
## [47] codetools_0.2-20 RcppRoll_0.3.1
## [49] tidyselect_1.2.1 UCSC.utils_1.4.0
## [51] farver_2.1.2 matrixStats_1.5.0
## [53] spatstat.explore_3.5-3 GenomicAlignments_1.44.0
## [55] jsonlite_2.0.0 progressr_0.16.0
## [57] ggridges_0.5.7 survival_3.8-3
## [59] systemfonts_1.3.1 tools_4.5.1
## [61] ragg_1.5.0 TFMPvalue_0.0.9
## [63] ica_1.0-3 Rcpp_1.1.0
## [65] glue_1.8.0 SparseArray_1.8.1
## [67] gridExtra_2.3 qs2_0.1.5
## [69] xfun_0.53 MatrixGenerics_1.20.0
## [71] dplyr_1.1.4 withr_3.0.2
## [73] fastmap_1.2.0 caTools_1.18.3
## [75] digest_0.6.37 R6_2.6.1
## [77] mime_0.13 textshaping_1.0.3
## [79] colorspace_2.1-2 scattermore_1.2
## [81] gtools_3.9.5 tensor_1.5.1
## [83] dichromat_2.0-0.1 spatstat.data_3.1-8
## [85] RSQLite_2.4.3 tidyr_1.3.1
## [87] data.table_1.17.8 S4Arrays_1.8.1
## [89] httr_1.4.7 htmlwidgets_1.6.4
## [91] uwot_0.2.3 pkgconfig_2.0.3
## [93] gtable_0.3.6 blob_1.2.4
## [95] lmtest_0.9-40 S7_0.2.0
## [97] htmltools_0.5.8.1 dotCall64_1.2
## [99] scales_1.4.0 Biobase_2.68.0
## [101] png_0.1-8 spatstat.univar_3.1-4
## [103] knitr_1.50 rjson_0.2.23
## [105] reshape2_1.4.4 curl_7.0.0
## [107] nlme_3.1-168 cachem_1.1.0
## [109] zoo_1.8-14 stringr_1.5.2
## [111] KernSmooth_2.23-26 parallel_4.5.1
## [113] miniUI_0.1.2 restfulr_0.0.16
## [115] desc_1.4.3 pillar_1.11.1
## [117] grid_4.5.1 vctrs_0.6.5
## [119] RANN_2.6.2 promises_1.3.3
## [121] stringfish_0.17.0 xtable_1.8-4
## [123] cluster_2.1.8.1 evaluate_1.0.5
## [125] cli_3.6.5 compiler_4.5.1
## [127] Rsamtools_2.24.1 rlang_1.1.6
## [129] crayon_1.5.3 future.apply_1.20.0
## [131] labeling_0.4.3 plyr_1.8.9
## [133] fs_1.6.6 stringi_1.8.7
## [135] viridisLite_0.4.2 deldir_2.0-4
## [137] BiocParallel_1.42.2 lazyeval_0.2.2
## [139] spatstat.geom_3.6-0 Matrix_1.7-4
## [141] RcppHNSW_0.6.0 patchwork_1.3.2
## [143] bit64_4.6.0-1 future_1.67.0
## [145] ggplot2_4.0.0 shiny_1.11.1
## [147] SummarizedExperiment_1.38.1 ROCR_1.0-11
## [149] igraph_2.1.4 memoise_2.0.1
## [151] RcppParallel_5.1.11-1 bslib_0.9.0
## [153] fastmatch_1.1-6 bit_4.6.0